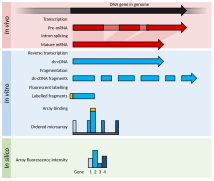
ᱨᱮᱫ:Summary of RNA-Seq.svg
ᱧᱮᱞᱚᱜ

Size of this PNG preview of this SVG file: ᱗᱖᱒ × ᱖᱐᱐ ᱯᱤᱠᱥᱮᱞ. ᱮᱴᱟᱜ ᱨᱤᱡᱚᱞᱩᱥᱚᱱᱥ: ᱓᱐᱕ × ᱒᱔᱐ ᱯᱤᱠᱥᱮᱞ | ᱖᱑᱐ × ᱔᱘᱐ ᱯᱤᱠᱥᱮᱞ | ᱙᱗᱖ × ᱗᱖᱘ ᱯᱤᱠᱥᱮᱞ | ᱑,᱒᱘᱐ × ᱑,᱐᱐᱘ ᱯᱤᱠᱥᱮᱞ | ᱒,᱕᱖᱐ × ᱒,᱐᱑᱕ ᱯᱤᱠᱥᱮᱞ | ᱑,᱐᱒᱐ × ᱘᱐᱓ ᱯᱤᱠᱥᱮᱞ
ᱟᱥᱚᱞ ᱨᱮᱫ (SVG ᱨᱮᱫ, ᱱᱚᱨᱢᱟᱞᱛᱮ ᱑,᱐᱒᱐ x ᱘᱐᱓ pixels, ᱨᱮᱫ ᱡᱟᱜᱟ: ᱒᱔᱖ KB)
ᱨᱮᱫ ᱨᱮᱭᱟᱜ ᱱᱟᱜᱟᱢ
ᱚᱠᱛᱚ ᱨᱮ ᱞᱤᱱ ᱢᱮ/ᱚᱠᱛᱚ ᱨᱮ ᱨᱮᱫ ᱧᱮᱞ ᱞᱟᱹᱜᱤᱛ ᱞᱤᱱ ᱢᱮ
| ᱢᱟᱹᱦᱤᱛ/ᱚᱠᱛᱚ | ᱴᱤᱯ | ᱡᱚᱠᱷᱟ | ᱵᱮᱵᱷᱟᱨᱤᱭᱟᱹ | ᱠᱟᱛᱷᱟ | |
|---|---|---|---|---|---|
| ᱱᱤᱛᱚᱜ | ᱑᱒:᱒᱐, ᱑᱓ ᱯᱷᱮᱵᱽᱨᱩᱣᱟᱨᱤ ᱒᱐᱑᱗ |  | ᱑,᱐᱒᱐ × ᱘᱐᱓ (᱒᱔᱖ KB) | Evolution and evolvability | text as paths to avoid artefacts |
| ᱑᱒:᱑᱙, ᱑᱓ ᱯᱷᱮᱵᱽᱨᱩᱣᱟᱨᱤ ᱒᱐᱑᱗ |  | ᱑,᱐᱒᱐ × ᱘᱐᱓ (᱙᱐ KB) | Evolution and evolvability | text as paths to avoid artefacts | |
| ᱑᱐:᱔᱘, ᱑᱓ ᱯᱷᱮᱵᱽᱨᱩᱣᱟᱨᱤ ᱒᱐᱑᱗ |  | ᱑,᱐᱒᱐ × ᱘᱐᱓ (᱘᱘ KB) | Evolution and evolvability | update sequence text to 'sans-serif' open font for more robust rendering | |
| ᱐᱘:᱔᱑, ᱑᱐ ᱯᱷᱮᱵᱽᱨᱩᱣᱟᱨᱤ ᱒᱐᱑᱗ |  | ᱑,᱐᱒᱐ × ᱘᱐᱓ (᱘᱗ KB) | Evolution and evolvability | reverse order of fragmentation and reverse-transcription | |
| ᱐᱕:᱐᱘, ᱕ ᱚᱠᱴᱚᱵᱚᱨ ᱒᱐᱑᱖ |  | ᱑,᱐᱒᱐ × ᱘᱐᱓ (᱘᱕ KB) | Evolution and evolvability | User created page with UploadWizard |
ᱯᱷᱟᱭᱤᱞ ᱵᱮᱣᱦᱟᱨ
ᱱᱚᱸᱰᱮ ᱫᱚ ᱟᱨ ᱮᱴᱟᱜ ᱥᱟᱦᱴᱟᱠᱚ ᱵᱟᱱᱩᱜ-ᱟ ᱡᱟᱸᱦᱟᱸ ᱥᱟᱶᱛᱮ ᱱᱤᱭᱟᱹ ᱨᱮᱫ ᱨᱮᱱᱟᱜ ᱡᱚᱱᱚᱲ ᱢᱮᱱᱟᱜ-ᱟ
ᱡᱮᱜᱮᱛ ᱡᱟᱠᱟᱛ ᱨᱮᱫ ᱵᱮᱵᱷᱟᱨᱟᱜ
ᱱᱚᱶᱟ ᱨᱮᱫᱠᱚ ᱵᱮᱵᱷᱟᱨᱟᱠᱟᱫ ᱣᱤᱠᱤᱠᱚ :
- ar.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- en.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- en.wikiversity.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- es.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- fa.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- gl.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- he.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- ja.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- ru.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ
- simple.wikipedia.org ᱨᱮ ᱵᱮᱣᱦᱟᱨ